 ABDPred: Antimicrobial Compound Prediction Server
ABDPred: Antimicrobial Compound Prediction Server What is an Antimicrobial compound/Antibiotics ?
As per the CDC website, Antimicrobial compounds are medicines that fight infections caused by bacteria in humans and animals by either killing the bacteria or making it difficult for the bacteria to grow and multiply
.
How bacteria become resistant to Antibiotics/Antimicrobial compounds ?
Antimicrobial compound resistance develops due to the following four major biochemical mechanisms: (i) Enzymatic inactivation or modification, (ii) Bypass
pathways, (iii) Altered target site, and (iv) Decreased uptake by (a) altering penetration or (b) via efflux pumps.
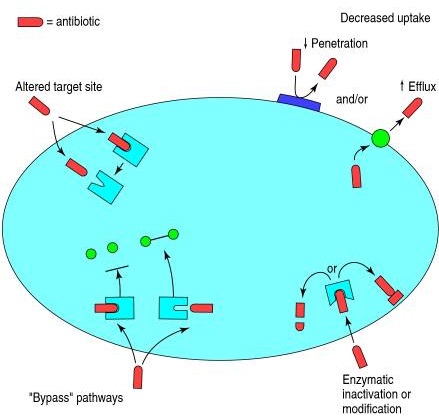
Figure: Four major biochemical mechanisms of Antimicrobial compound resistance. Source: Hawkey, P. M. (1998). The origins and molecular basis of Antimicrobial compound resistance. BMJ, 317(7159), 657–660. https://doi.org/10.1136/bmj.317.7159.657
What is ABDPred ?
ABDPred is a prediction server that can predict Antimicrobial compound drugs using four machine learning (ML) classifiers: XGBoost (XGB), Random Forest (RF), Gradient Boosting (GBC), and Deep Neural Network (DNN), individually, and aggregately using the soft voting technique. The server is able to predict whether the input chemical/drug might act as an Antimicrobial compound drug. The ‘druglikeness and bioavailability profile’ of the input chemical/drug is also checked in the server. In addition, the server is able to compare the two-dimensional (2D) structure of input drugs to known Antimicrobial compound drugs (n=390) based on the Tanimoto Coefficient Similarity score..
How does the ABDpred server work ?
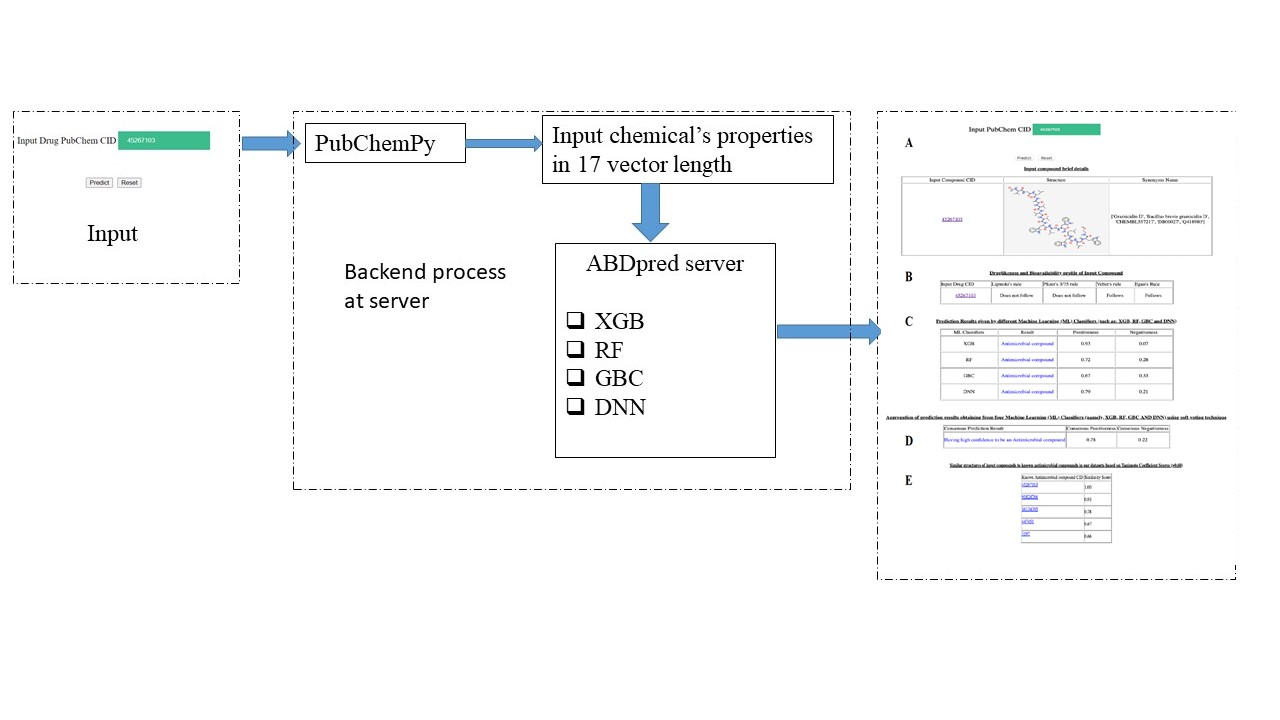
Figure. ABDpred performs prediction after users input as drug PubChem CID value
How are the Consensus Prediction Result and scores (positiveness and negativeness) generated ?
In ABDpred server, the four machine learning (ML) classifers, such as XGB (XGBoost), RF (Random Forest), GBC (Gradient Boosting) and DNN (Deep Neural Network) have been used to classify Antimicrobial compound drugs. The prediction score of each of the four ML classifier models is described using positiveness and negativeness. The Positiveness score is calculated using predict_proba(Z)[:1] and the negative score is calculated using predict_proba(Z)[:0].
| Prediction score (Positiveness) | Prediction Result |
| If PositivenessXGB >=0.50 | Result: Antimicrobial compound Drug otherwise Not Antimicrobial compound Drug |
| If PositivenessRF >=0.50 | Result: Antimicrobial compound Drug otherwise Not Antimicrobial compound Drug |
| If PositivenessGBC >=0.50 | Result: Antimicrobial compound Drug otherwise Not Antimicrobial compound Drug |
| If PositivenessDNN >=0.55 | Result: Antimicrobial compound Drug otherwise Not Antimicrobial compound Drug |
Consensus Prediction result and scores using soft voting technique:
| Bins | Consensus Prediction score (Positiveness) | Prediction Result |
| 0.70 to 1.0 | x̄>=0.70 | Having high confidence to be an Antimicrobial compound |
| 0.60 to 0.70 | 0.60>x̄>=0.70 | Medium confidence to be an Antimicrobial compound |
| 0.50 to 0.60 | 0.50>=x̄>=0.60 | Low confidence to be an Antimicrobial compound |
| 0.0 to 0.50 | x̄<0.50 AND PositivenessXGB < 0.50 AND PositivenessRF < 0.50 AND PositivenessGBC < 0.50 AND PositivenessDNN < 0.50 | Having high Confidence to be a Non-Antimicrobial compound |
| 0.0 to 0.50 | x̄<0.50 AND PositivenessXGB>=0.55 | Having High Confidence to be a Non-Antimicrobial compound. However, XGBoost algorithm alone predicts it as an Antimicrobial compound. Kindly check the structure and physicochemical properties. |
| 0.0 to 0.50 | x̄<0.50 AND PositivenessRF>=0.55 | Having High Confidence to be a Non-Antimicrobial compound. However, Random Forest algorithm alone predicts it as an Antimicrobial compound. Kindly check the structure and physicochemical properties. |
| 0.0 to 0.50 | x̄<0.50 AND PositivenessGBC>=0.55 | Having High Confidence to be a Non-Antimicrobial compound. However, Gradient Boosting algorithm alone predicts it as an Antimicrobial compound. Kindly check the structure and physicochemical properties. |
| 0.0 to 0.50 | x̄<0.50 AND PositivenessDNN>=0.55 | Having High Confidence to be a Non-Antimicrobial compound. However, Deep Neural Network algorithm alone predicts it as an Antimicrobial compound. Kindly check the structure and physicochemical properties. |
Can user(s) download the datasets and source codes of ABDPred ?
User can download the datasets and sources codes of ABDpred server developement by visiting the download page
What are the limitations of the ABDPred server ?
If the input chemical/drug does not have any XLogP3-AA value record in the PubChem database, then the ABDPred server might be unable to predict properly as an Antimicrobial compound drug. In that case, please check the other physicochemical properties and relevant literature for the input drug.
How to calculate and compare the Tanimoto coffecient score of input drug against 390 known Antimicrobial compound drugs ?
First, the input drug is coverted into PubChem fingerprint. The fingerprint of 390 known Antimicrobial compound drugs has been computed previously. Then comparison betwen input drug and known Antimicrobial compound drug 1. The process is continued till the 389 times.
The program pseudo code:
def tanimoto(compound1, d):
fp1 = int(compound1.fingerprint, 16)
fp2 = int(d, 16)
fp1_count = bin(fp1).count('1')
fp2_count = bin(fp2).count('1')
both_count = bin(fp1 & fp2).count('1')
return ((both_count) / (fp1_count + fp2_count - both_count))
How to evaluate druglikeness and bioavailability profile of the input chemical/drug ?
The molecular weight (mw), hydrogenbond donor count (hbdc), hydrogenbond acceptor count (Hdac) and partition coefficient (xlogp3), topological surface area (tpsa) and rotatable bond count (rbc) of input drug is curated from PubChemPy python tool. Then we computed the conditions of four rules that druglikeness and bioavailability profile of input drug.
Such as:
i) Lipinski's rule: mw < 500 and hbdc <= 5 and hdac <=10 and xlogp3 <=5
ii) Pfizer 3/75: xlogp3 >=3 and tpsa <=75
iii) Veber's rule: tpsa >=140 and rbc >=10
iv) Egan's Rule: tpsa >=131.6 and xlogp3 >=5.88